Examples
Several user examples are available directly in Geant4 for the usage of Geant4-DNA processes and models. They available in both extended and advanced categories of Geant4 examples.
They cover Geant4-DNA physics (track structures), chemistry (radiolysis simulation), various geometries of biological targets and simulation of early DNA damage from direct and indirect effects.
These examples are located in the $G4INSTALL/examples directory of the Geant4 installation. A README file is provided with each example directly in their source directory.
Their online documentation is included in Geant4.
A dedicated Geant4 forum is available here.
Physics
- The « clustering » extended/medical/dna example illustrates how to identify ionisation clusters.
- The « dnaphysics » extended/medical/dna example shows how to simulate track structures in liquid water using the Geant4-DNA physics processes and models, assembled in Geant4-DNA physics constructors. It also explains how to extract physical information at the step and track levels, such as process type, position, energy deposited, scattering angle... and how to use the variable density material feature for liquid water.
- The « icsd » extended/medical/dna example illustrates how to use cross section models for DNA-related materials.
- The « jetcounter » extended/medical/dna example explains how to simulate a gas dosimeter.
- The « microdosimetry » extended/medical/dna example shows how to use Geant4 and Geant4-DNA physics models in different regions of the geometrical setup, using the G4EmDNAPhysicsActivator class.
- The « microprox » extended/medical/dna example explains how to simulate microdosimetry proximity functions.
- The « microtrack » extended/medical/dna example explains how to simulate microdosimetry quantities for ions.
- The « microyz » extended/medical/dna example explains how to simulate microdosimetry quantities for electrons.
- The « mfp » extended/medical/dna example explains how to extract mean free paths.
- The « phasespace » extended/medical/dna example explains how to produce phase space files from Geant4 physics processes and models.
- The « radial » extended/medical/dna example explains how to simulate radial doses around ion tracks.
- The « range » extended/medical/dna example explains how to simulate track length, projected range and penetration of particles.
- The « slowing » extended/medical/dna example explains how to simulate slowing down spectra.
- The « splitting » extended/medical/dna example explains how to accelerate simulation through splitting in ionisation.
- The « spower » extended/medical/dna example explains how to simulate stopping powers.
- The « svalue » extended/medical/dna example explains how to simulate "S-values" in spherical targets of liquid water.
- The « wvalue » extended/medical/dna example explains how to simulate "W-values" in liquid water.
- The « AuNP » extended/medical/dna example explains how to simulate track structures of electrons in a microscopic gold volume immersed in liquid water.
- The « TestEm5 » extended/electromagnetic example explains how to extract atomic deexcitation information (using the dna.mac macro file).
- The « TestEm12 » extended/electromagnetic example explains how to extract radial dose distributions in spherical shells of liquid water (such as dose point kernels, using the dna.mac macro file).
Chemistry
- The « chem1 » extended/medical/dna example illustrates how to activate the simulation of water radiolysis (step-by-step method).
- The « chem2 » extended/medical/dna example illustrates how to set minimum time step limits on water radiolysis (step-by-step method).
- The « chem3 » extended/medical/dna example illustrates how to implement user actions in the chemistry module (step-by-step method).
- The « chem4 » extended/medical/dna example illustrates how to compute radiochemical yields ("G") versus time, including a dedicated ROOT graphical interface (step-by-step method).
- The « chem5 » extended/medical/dna example illustrates how to compute radiochemical yields ("G") versus time, using alternative physics and chemistry lists (step-by-step method).
- The « chem6 » extended/medical/dna example illustrates how to compute radiochemical yields ("G") versus time and LET using IRT method.
- The « molcounters » extended/medical/dna examples illustrate how to score species in geometries.
- The « scavenger » extended/medical/dna example illustrates how to simulate scavenging using an easy-to-use interface and the IRT method.
- The « UHDR » extended/medical/dna example illustrates how to activate the chemistry mesoscopic model in combination with the step-by-step model, and allows to simulate chemical reactions beyond 1 us post-irradiation.
Geometry
- The Geant4 « microbeam » advanced example explains how to define a realistic cellular phantom based on confocal microscopy imaging for the simulation of cellular irradiation in single ion mode, based on the LP2iB-CENBG microbeam irradiation facility.
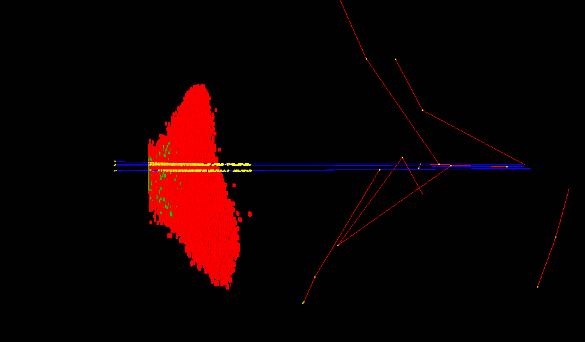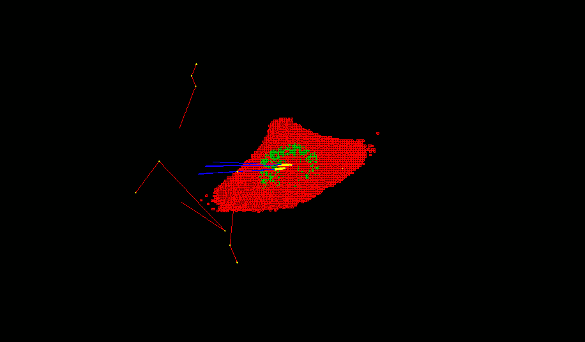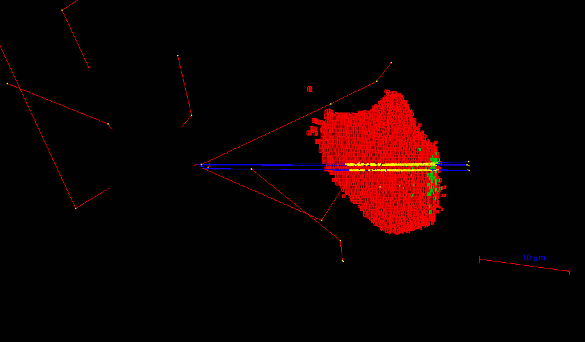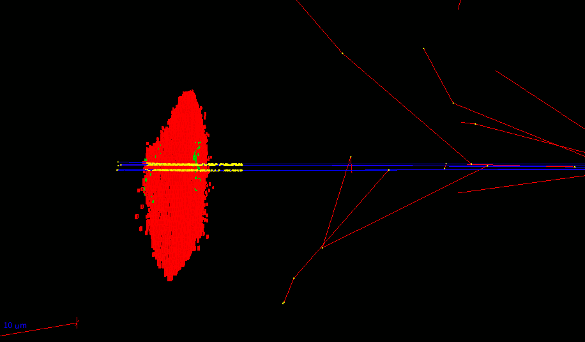
- The Geant4 « cellularPhantom » advanced/dna example explains how to define a realistic cellular population phantom based on confocal microscopy imaging. It can be used for dosimetry and it is further described here.
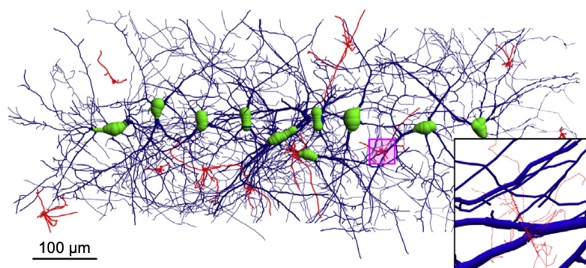
- The Geant4 « neuron » extended/medical/dna example shows how to simulate a neural network including physics and radiolysis.
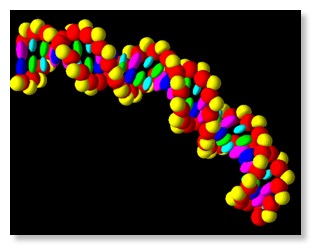
- The Geant4 « pdb4dna » extended/medical/dna example shows how to model molecular geometries from the Protein Data Bank (PDB) and record energy deposition events in biological volumes of interest. It is further described at a dedicated web site: http://pdb4dna.in2p3.fr.

- The Geant4 « wholeNuclearDNA » extended/medical/dna example shows how to model a simplified cellular nucleus containing a geometrical model for the DNA molecule.
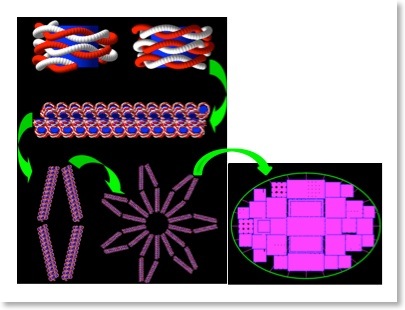
- A short guide on how DNA geometries may be modelled in Geant4 is available here, written by Nathanael Lampe.
Biological damage
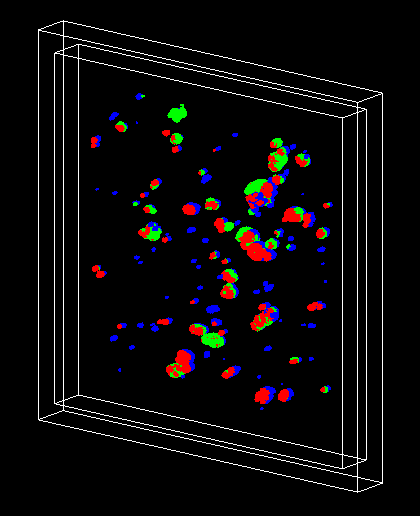
- The Geant4 « dnadamage1 » extended/medical/dna example shows how to simulate early DNA damage (direct + indirect) in a cubic voxel of 40 nm side containing a piece of heterochromatin generated with the DNAFabric software (link).
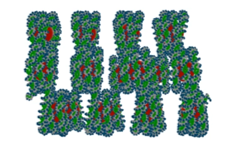
- The Geant4 « dnadamage2 » extended/medical/dna example provides scoring of plasmid DNA strand breaks using the IRT method. It extends the chem6 example by adding DNA molecule information and the scoring of Strand Breaks. Experimental conditions are considered; such as oxygen and DMSO molar concentrations.
- The Geant4 « dsbandrepair » advanced/dna example shows how to simulate direct and indirect DNA damage in various geometries of biological targets.
- The Geant4 « moleculardna » advanced/dna example shows how to simulate direct and indirect DNA damage in various geometries (including plasmids, E. coli bacterium and various human cells).

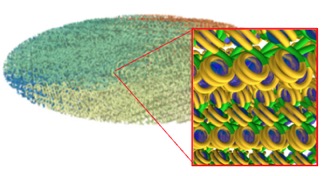
It is further described at a dedicated web site:
http://moleculardna.org
